Overview
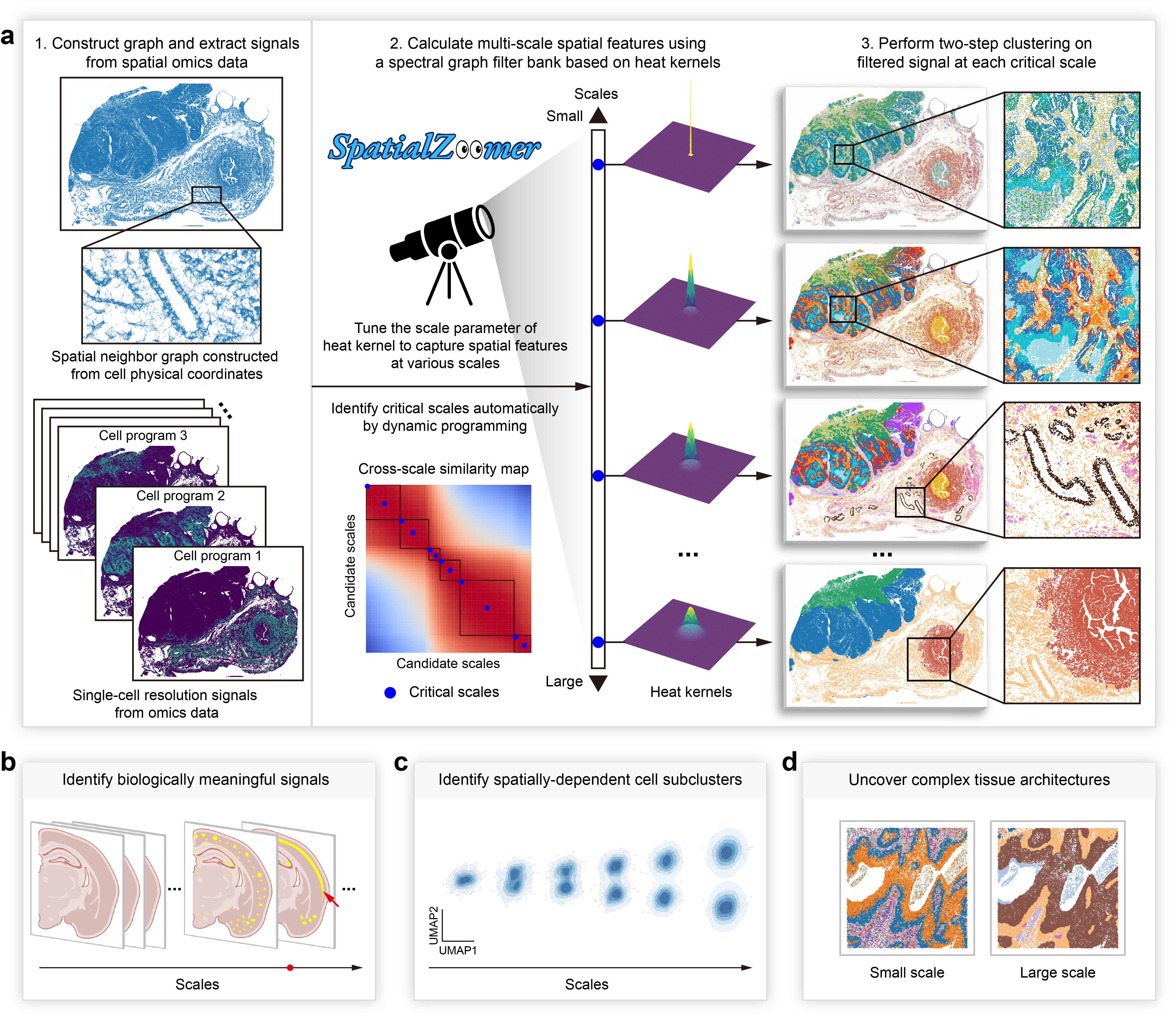
SpatialZoomer is a novel computational framework for multi-scale feature analysis of single-cell resolved spatial transcriptomics, functioning like a zoom-capable microscope that captures spatial structures from cells, niches, to tissue domains. It further identifies critical scales automatically by partitioning the cross-scale similarity map via dynamic programming.
SpatialZoomer supports diverse downstream applications, such as identifying biologically meaningful signals, spatially dependent cell subclusters, and uncovering complex tissue architectures.
SpatialZoomer scales to datasets with millions of cells with high computational efficiency and low hardware requirements.
Demo
Note: Across scales, cluster colors are aligned by an algorithm that maximizes the overlap between clusters.
Please slide the scale axis to explore the results at critical scales (higher values result in more smoothed signals, identifying more global spatial structures):
Please choose the clustering resolution (a higher value results in more clusters):
Installation
# Create and activate a virtual environment
conda create -n spatialzoomer python=3.10 -y
conda activate spatialzoomer
# Install required dependency
conda install -c conda-forge pyarrow
# Install SpatialZoomer
pip install SpatialZoomer==1.0.0
Tutorial
To get started with SpatialZoomer, please refer to our tutorials available in the GiHub repository.